Emergence
and Diffusion
of Multiple Drug Resistance
against antibiotics (EDIRA)


Our team mainly aims to elucidate the mechanisms of emergence and diffusion of multiple antimicrobial resistance (AMR) focusing on the Multi Drug Resistance (MDR) Mycobacteria, including Mycobacterium tuberculosis (Mtb) and MDR Enterobacteriaceae, especially Klebsiella pneumoniae species.
The team also uses several specific infection models to evaluate the in vivo activity of new antibiotic- or non-antibiotic (such as bacteriophages) -based strategies able to treat mycobacteria infections, or prevent / suppress colonisation by AMR Enterobacteriaceae.
Furthermore, the team also investigates neglected infectious diseases such as Buruli Ulcer, leprosy and rhinoscleroma (e.g. Klebsiella pneumoniae subspecies pathogenesis). Moreover, the team runs also the French National Reference Centre (NRC) of Mycobacteria.

The team members
Our research focus on 4 axes
- Mycobacteria
- Enterobacteriaceae
- Antimicrobial resistance
- Epidemiology
- Neglected Infectious diseases
- Bacteriophage
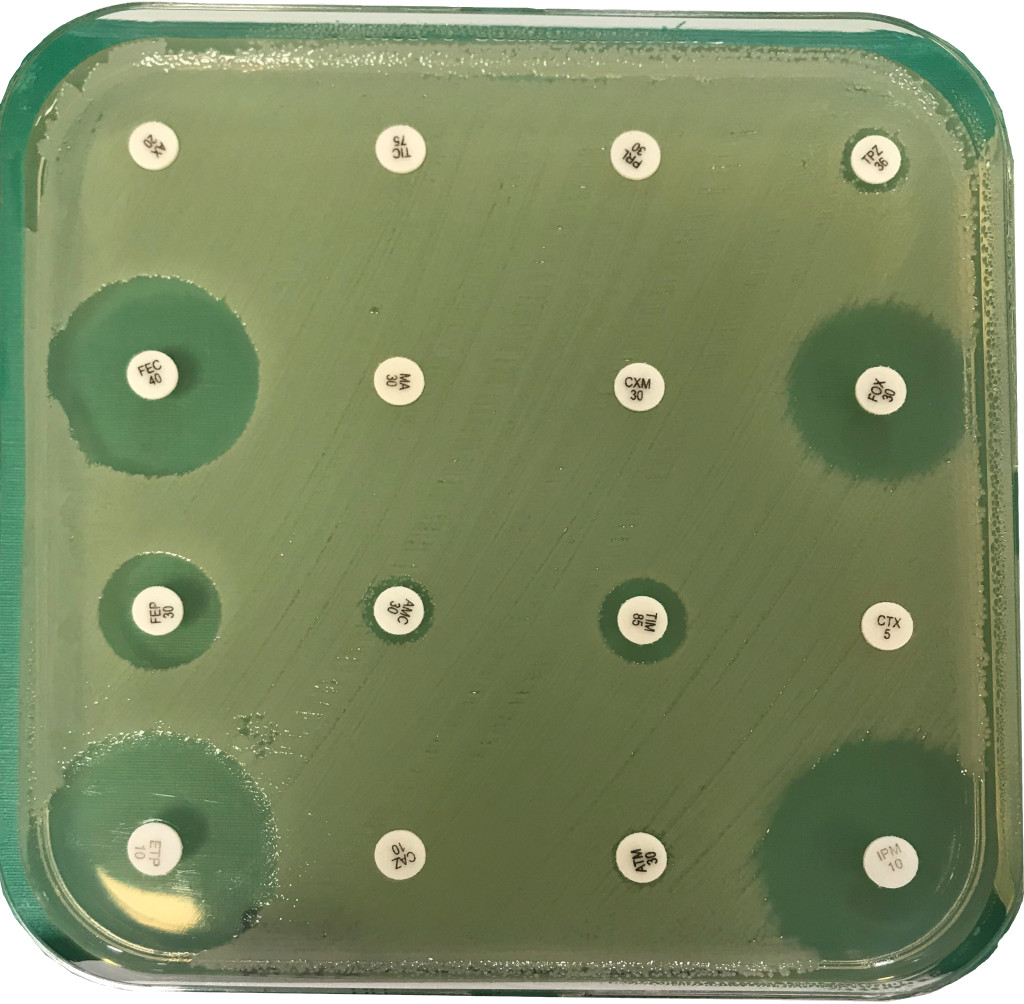
1. Epidemiology of antimicrobial resistance (AMR) (Enterobacteriaceae and mycobacteria)
A better knowledge of the prevalence, risk factors, and molecular epidemiology of MDR bacteria is of utmost public health importance.
Our research is based on the participation to and/or coordination of national and international surveillance and research networks or cohorts (ONERBA , National Reference Center for Mycobacteria , AZAY-Mycobacteria , group mycobacteria of the Fench Society for Microbiology, TBnet , ESGMYC , ECDC and WHO ) in collaboration with clinicians and national experts.
These collaborations allowed us to better assess the effectiveness of new drugs such as bedaquiline for MDR-TB, and to analyse the epidemiology of MDR-TB in France, Europe or Asia and the epidemiology of MDR Enterobacteriaceae in France (ESBL or carbapenemase-positive) or abroad (China, Vietnam).
2. Novel strategies to fight antimicrobial resistance (AMR) in Enterobacteriaceae and mycobacteria
Because Enterobacteriaceae infections (e.g Klebsiella pneumoniae (KP)) mainly result from prior colonization, we are trying to better understand the capacity of KP to colonize the gut microbiota and to identify new approaches (e.g. bacteriophages) to specifically eliminate MDR KP from the gut.
We are also developing new antibiotics and antibiotics regimens that are evaluated for their efficacy using preclinical models of mycobacteria infections (M. tuberculosis, M. avium and M. abscessus) for which we have extensive and recognized expertise.
Moreover, we are developing experimental evolution approaches to identify new mechanisms directly involved in or facilitating AMR emergence, as well as the host impact on emergence of AMR.
3. Study of molecular mechanisms of drug activity and resistance in mycobacteria
Multidrug resistant (MDR) tuberculosis (TB) bacteria being increasingly isolated, our goals are to (i) investigate the molecular mechanisms accounting for drug resistance in tuberculosis, (ii) provide a comprehensive assessment of MDR tuberculosis in France, (iii) improve drug-susceptibility testing and in silico predictions, and (iv) target the genetic basis underlying the spread and success of major MDR M. tuberculosis lineages.
Capitalizing on 1,300 MDR isolates longitudinally collected in France, we combine active surveillance data with advanced genomic epidemiology and phylogenetic analysis to identify mutations involved in antimicrobials resistance and the transmission dynamics of TB. Because M. tuberculosis is a slowly growing bacterium, we expect to develop faster AMR prediction tools that are essential for prescription of personalised TB patients’ treatments.
4. Neglected infectious diseases such as Buruli Ulcer, leprosy and rhinoscleroma
We are involved in studies evaluating new strategies to treat neglected infectious diseases such as Buruli Ulcer, and leprosy.
We also investigate how K. rhinoscleromatis, causing a chronic disease, and K. pneumoniae, causing acute infections, differently interact with and manipulate the host response during respiratory infections, focusing especially on their interaction with monocytes.
The opportunities
- to provide novel insights into new antimicrobial drug targets
- to provide new insights in antibacterial drug resistance
- to develop allele recombination-based approaches to decipher mechanisms involved in transmission of tuberculosis thorough multidisciplinary approaches and consortium involving clinicians, microbiologists, bioinformaticians and epidemiologists, as well as pharma or European initiatives
Publications
Oral Regimens for Rifampin-Resistant, Fluoroquinolone-Susceptible Tuberculosis. Guglielmetti LN Engl J Med. 2025 Jan 30;392(5):468-482. https://doi: 10.1056/NEJMoa2400327
Bedaquiline, delamanid, linezolid, and clofazimine for rifampicin-resistant and fluoroquinolone-resistant tuberculosis (endTB-Q): an open-label, multicentre, stratified, non-inferiority, randomised, controlled, phase 3 trial. Lancet Respir Med. 2025 Sep;13(9):809-820. https://doi: 10.1016/S2213-2600(25)00194-8
Bedaquiline Activity against Leprosy. N Engl J Med. 2025 Jun 5;392(21):2174-2176. https://doi.org/10.1056/nejmc2412487
Evaluation of the role of whiB6 and kdpDE in the dominant multidrug-resistant clone Mycobacterium tuberculosis B0/W148. Bonnet I, Orgeur M, Brossier F, Sayes F, Frigui W, Madacki J, Varet H, Chauffour A, Aubry A, Veziris N, Sougakoff W, Brosch R, Tournebize R. Microbiol Spectr. 2025. 13:e0322424. https://doi.org/10.1128/spectrum.03224-24
Oral Regimens for Rifampin-Resistant, Fluoroquinolone-Susceptible Tuberculosis. Guglielmetti L, Khan U, Velásquez GE, Gouillou M, Abubakirov A, Baudin E, Berikova E, Berry C, Bonnet M, Cellamare M, Chavan V, Cox V, Dakenova Z, de Jong BC, Ferlazzo G, Karabayev A, Kirakosyan O, Kiria N, Kunda M, Lachenal N, Lecca L, McIlleron H, Motta I, Toscano SM, Mushtaque H, Nahid P, Oyewusi L, Panda S, Patil S, Phillips PPJ, Ruiz J, Salahuddin N, Garavito ES, Seung KJ, Ticona E, Trippa L, Vasquez DEV, Wasserman S, Rich ML, Varaine F, Mitnick CD; endTB Clinical Trial Team. N Engl J Med. 2025. 392:468-482. https://doi.org/10.1056/nejmoa2400327
Characterization of the Orphan Cytochrome P450 CYP135B1 from Mycobacterium tuberculosis: Involvement in Metabolism but Not in the Antibacterial Activity of the Antitubercular Drug SQ109. Sadowski E, Pietrancosta N, Veyron-Churlet R, Boucher JL, Pionneau C, Clodic G, Matheron L, Poch O, Mayer C, Sachon E, Aubry A. ACS Infect Dis. 2025 ; 11:869-881. https://doi.org/10.1021/acsinfecdis.4c00893
Levofloxacin activity at increasing doses in a murine model of fluoroquinolone-susceptible and -resistant tuberculosis. Maitre T, Godmer A, Mory C, Chauffour A, Mai TC, El Helali N, Aubry A, Veziris N. Antimicrob Agents Chemother. 2024; 68:e0058324. https://doi.org/10.1128/aac.00583-24
Contribution of MALDI-TOF mass spectrometry and machine learning including deep learning techniques for the detection of virulence factors of Clostridioides difficile strains. Godmer A, Giai Gianetto Q, Le Neindre K, Latapy V, Bastide M, Ehmig M, Lalande V, Veziris N, Aubry A, Barbut F, Eckert C. Microb Biotechnol. 2024 ; 17:e14478. https://doi.org/10.1111/1751-7915.14478
Overall Structures of Mycobacterium tuberculosis DNA Gyrase Reveal the Role of a Corynebacteriales GyrB-Specific Insert in ATPase Activity. Petrella S, Capton E, Raynal B, Giffard C, Thureau A, Bonneté F, Alzari PM, Aubry A*, Mayer C* (*co-last authors). Structure, 2019; 27:579-589.e5. https://doi.org/10.1016/j.str.2019.01.004
Fully weekly antituberculosis regimen: a proof-of-concept study. Kort F, Fournier Le Ray L, Chauffour A, Jarlier V, Lounis N, Andries K, Aubry A, Guglielmetti L, Veziris N (2020). Eur Respir J 56:1902502 .
https://doi.org/10.1183/13993003.02502-2019